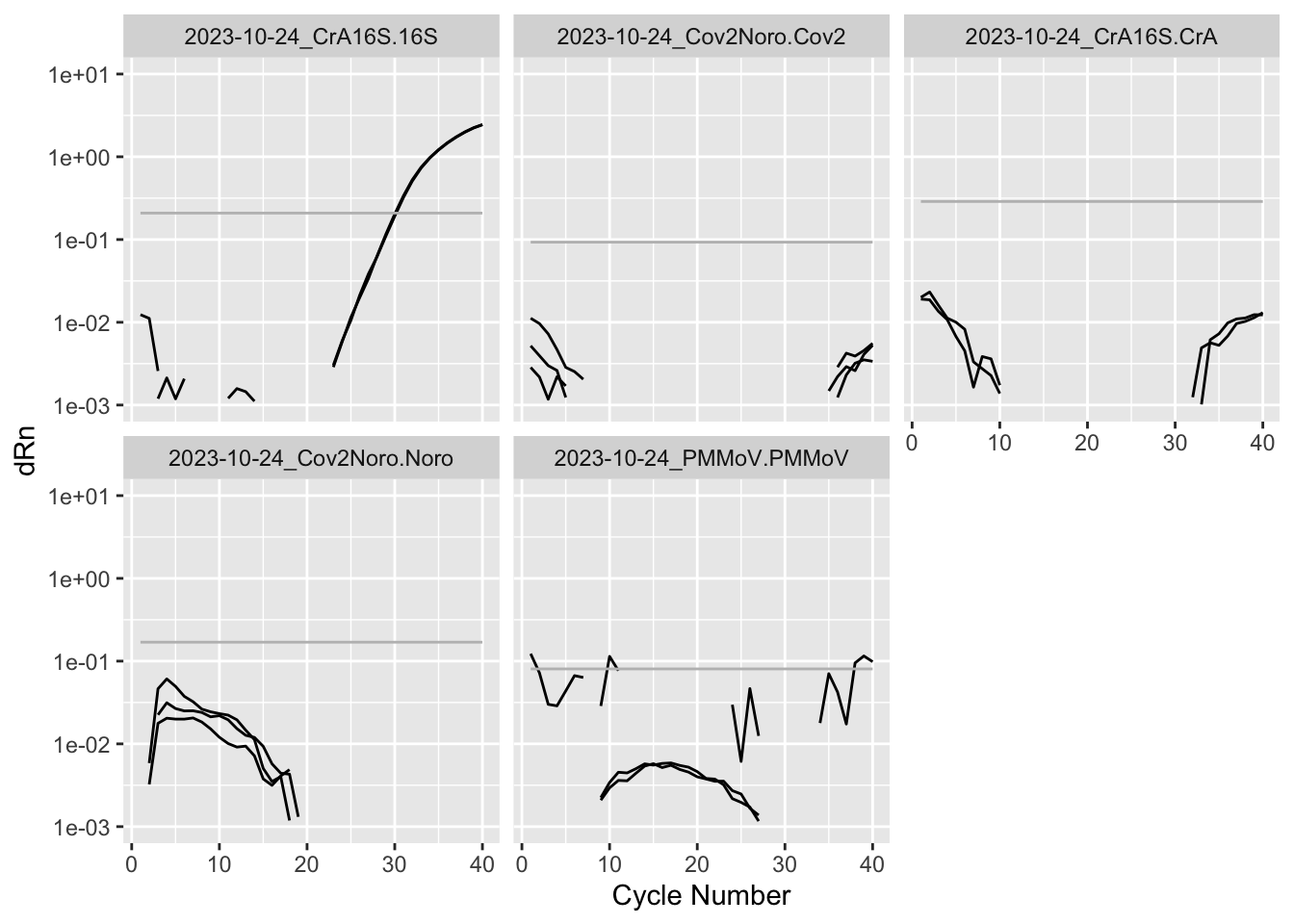
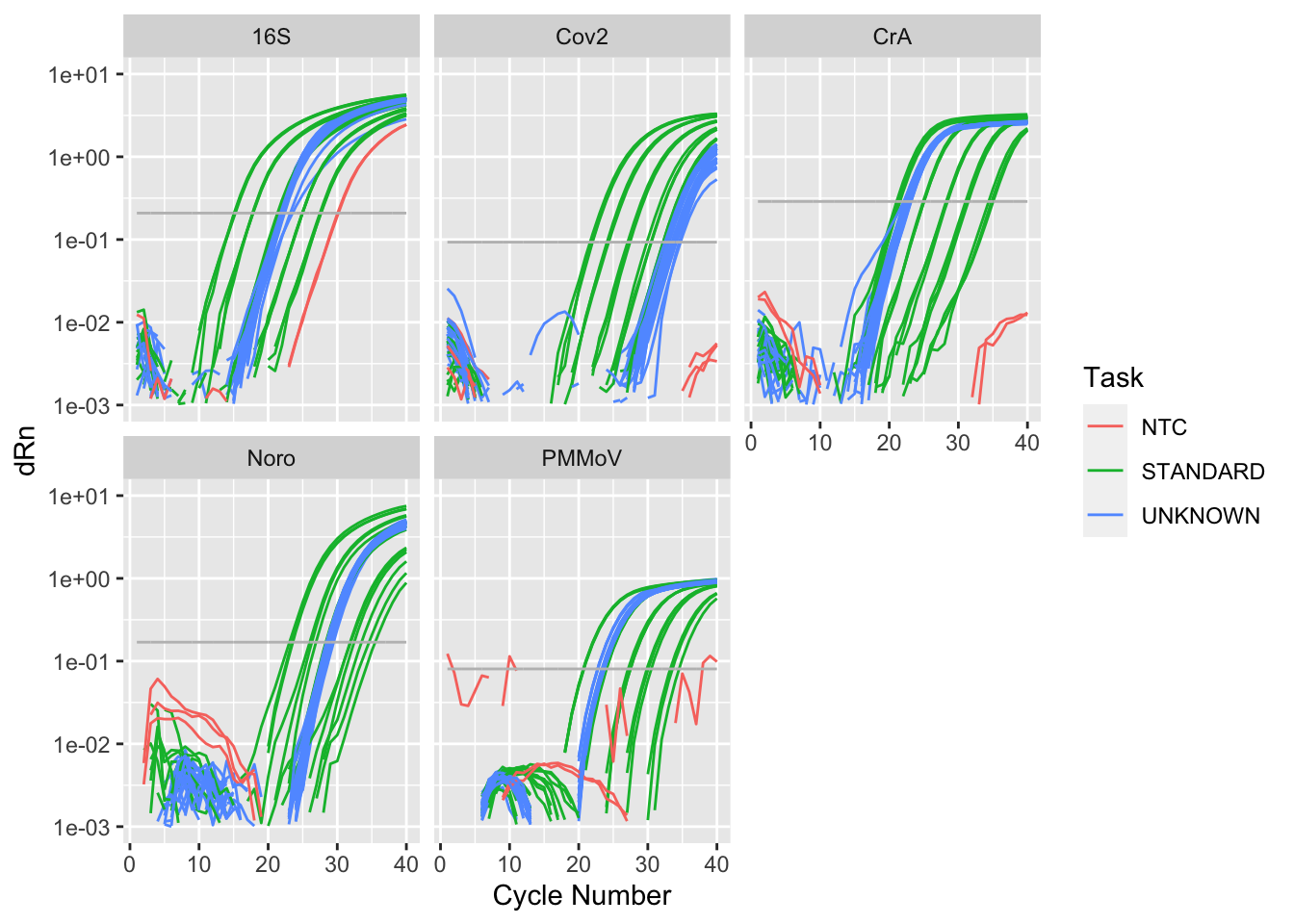
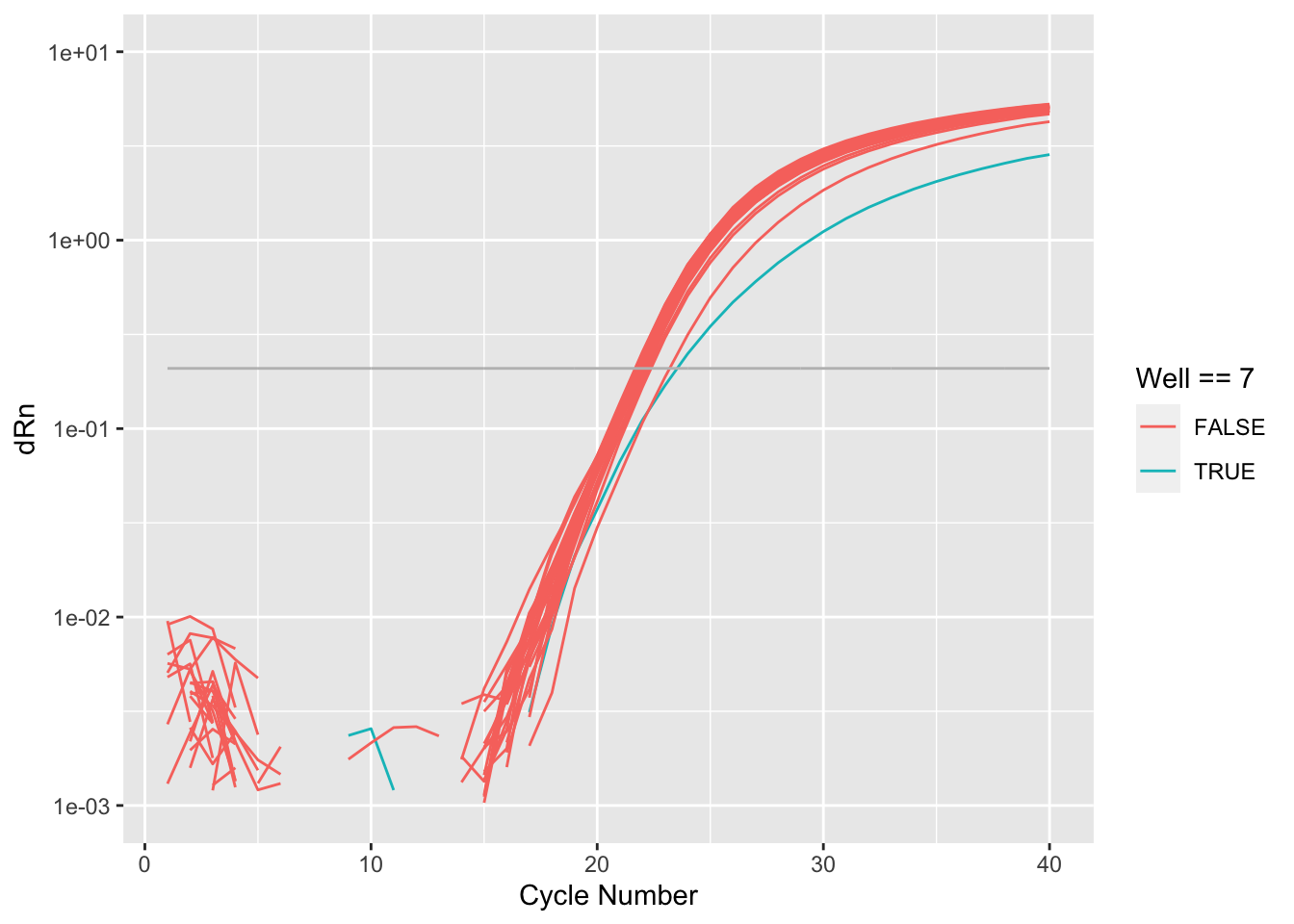
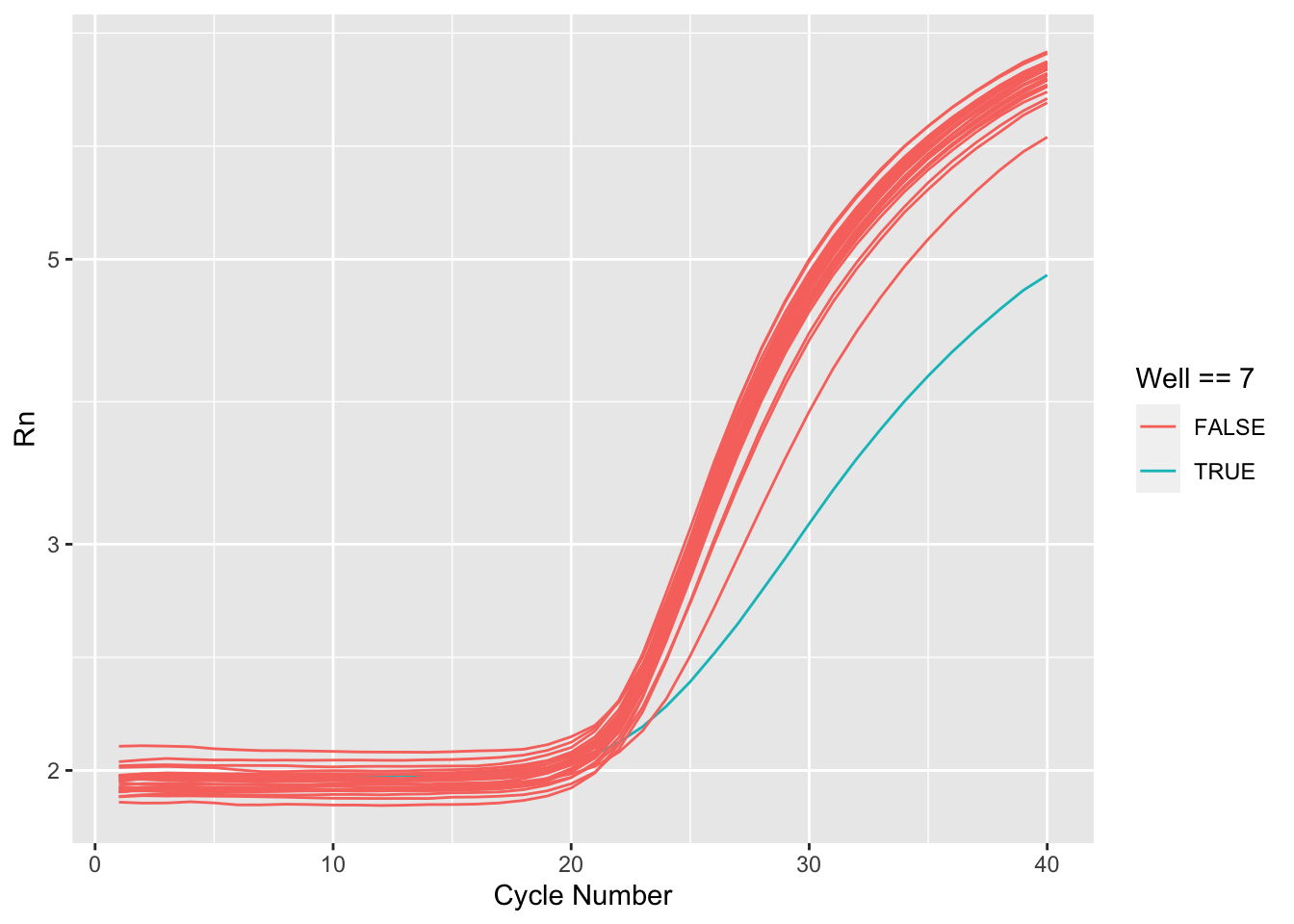
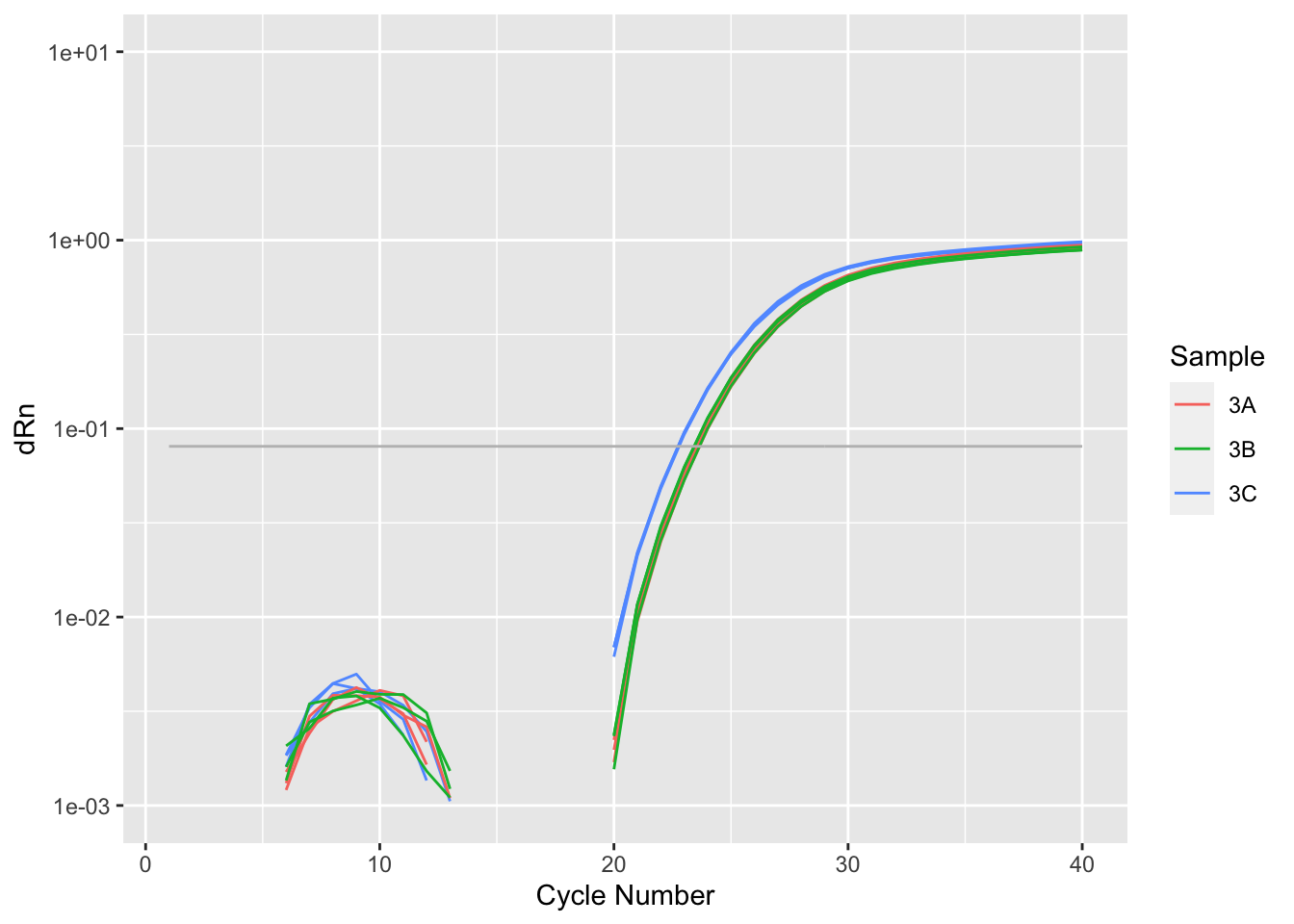
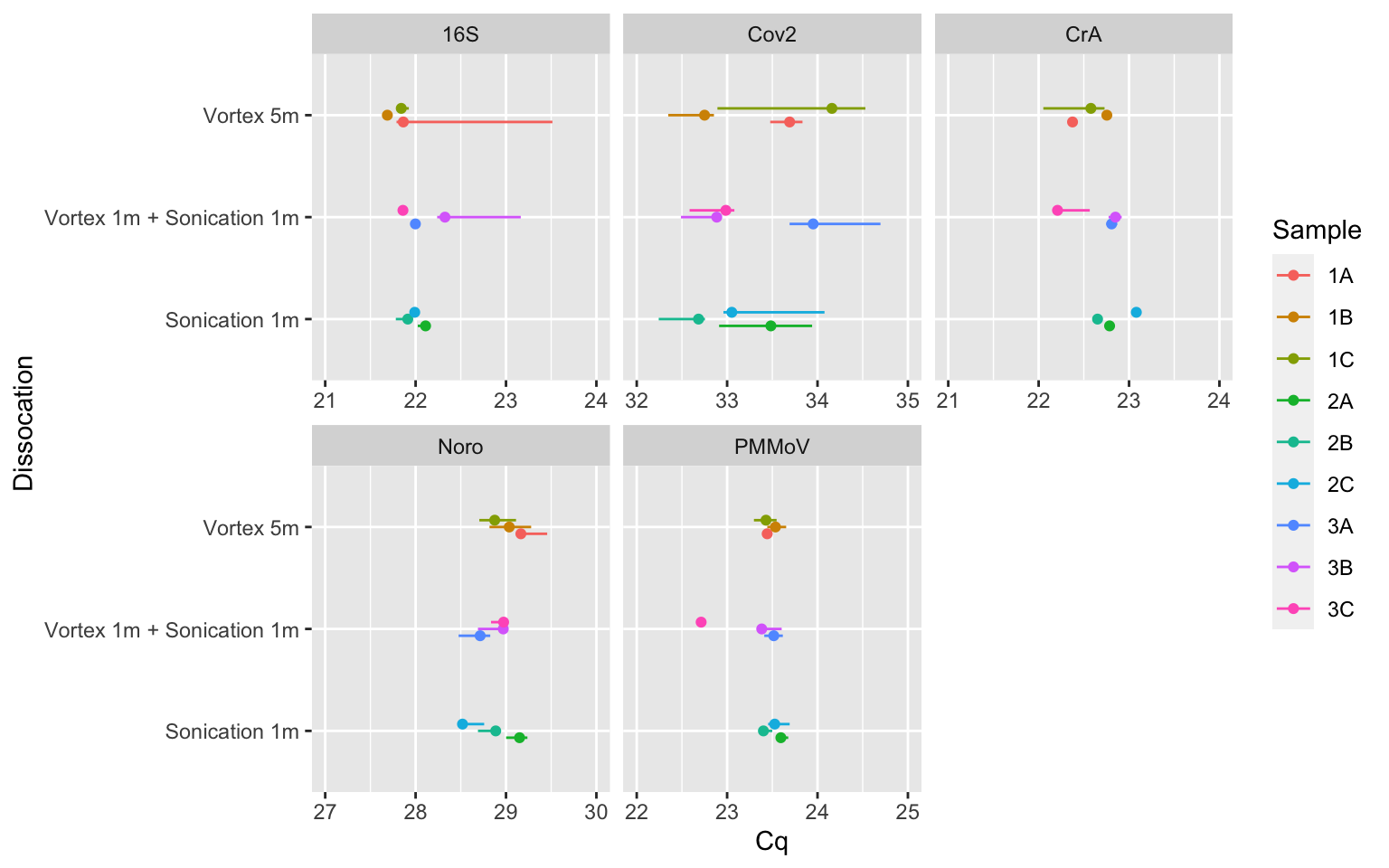
Rows: 8,920
Columns: 38
$ Well <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ `Well Position` <chr> "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1"…
$ `Cycle Number` <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,…
$ Target <chr> "Cov2", "Cov2", "Cov2", "Cov2", "Cov2", "Cov2"…
$ Rn <dbl> 0.4810576, 0.4760514, 0.4685638, 0.4619383, 0.…
$ dRn <dbl> 0.0255262889, 0.0208546813, 0.0137017026, 0.00…
$ Sample <chr> "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A"…
$ Omit <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALS…
$ plate <chr> "2023-10-24_Cov2Noro", "2023-10-24_Cov2Noro", …
$ well_row <chr> "A", "A", "A", "A", "A", "A", "A", "A", "A", "…
$ well_col <chr> "1", "1", "1", "1", "1", "1", "1", "1", "1", "…
$ Task <chr> "UNKNOWN", "UNKNOWN", "UNKNOWN", "UNKNOWN", "U…
$ Reporter <chr> "FAM", "FAM", "FAM", "FAM", "FAM", "FAM", "FAM…
$ Quencher <chr> "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "N…
$ `Amp Status` <chr> "AMP", "AMP", "AMP", "AMP", "AMP", "AMP", "AMP…
$ `Amp Score` <dbl> 1.285299, 1.285299, 1.285299, 1.285299, 1.2852…
$ `Curve Quality` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ `Result Quality Issues` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ Cq <dbl> 33.47677, 33.47677, 33.47677, 33.47677, 33.476…
$ `Cq Confidence` <dbl> 0.9706938, 0.9706938, 0.9706938, 0.9706938, 0.…
$ `Cq Mean` <dbl> 33.66743, 33.66743, 33.66743, 33.66743, 33.667…
$ `Cq SD` <dbl> 0.1797476, 0.1797476, 0.1797476, 0.1797476, 0.…
$ `Auto Threshold` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ Threshold <dbl> 0.09301463, 0.09301463, 0.09301463, 0.09301463…
$ `Auto Baseline` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ `Baseline Start` <dbl> 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
$ `Baseline End` <dbl> 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29, 29…
$ `Treatment Group` <chr> "1", "1", "1", "1", "1", "1", "1", "1", "1", "…
$ Dissocation <chr> "Vortex 5m", "Vortex 5m", "Vortex 5m", "Vortex…
$ `Vortex time (min)` <dbl> 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5…
$ `Sonication time (min)` <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ CollectionDate <date> 2023-10-17, 2023-10-17, 2023-10-17, 2023-10-1…
$ Source <chr> "Sludge", "Sludge", "Sludge", "Sludge", "Sludg…
$ `Filtration time (sec)` <dbl> 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10…
$ `CP Time` <time> 03:37:00, 03:37:00, 03:37:00, 03:37:00, 03:37…
$ `Shield Added (uL)` <dbl> 400, 400, 400, 400, 400, 400, 400, 400, 400, 4…
$ `Qubit RNA` <dbl> 24.2, 24.2, 24.2, 24.2, 24.2, 24.2, 24.2, 24.2…
$ `Qubit DNA` <chr> "40", "40", "40", "40", "40", "40", "40", "40"…