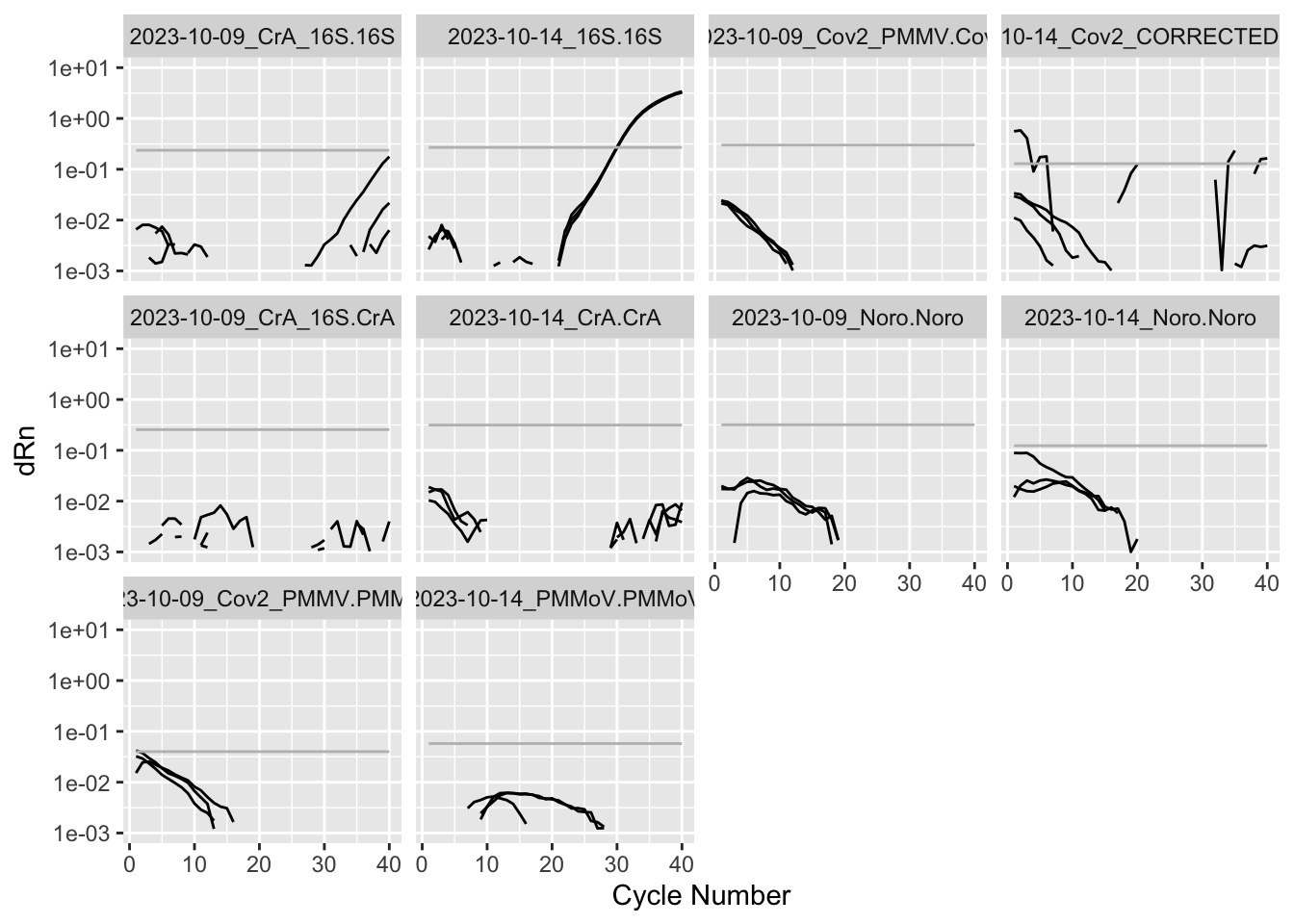
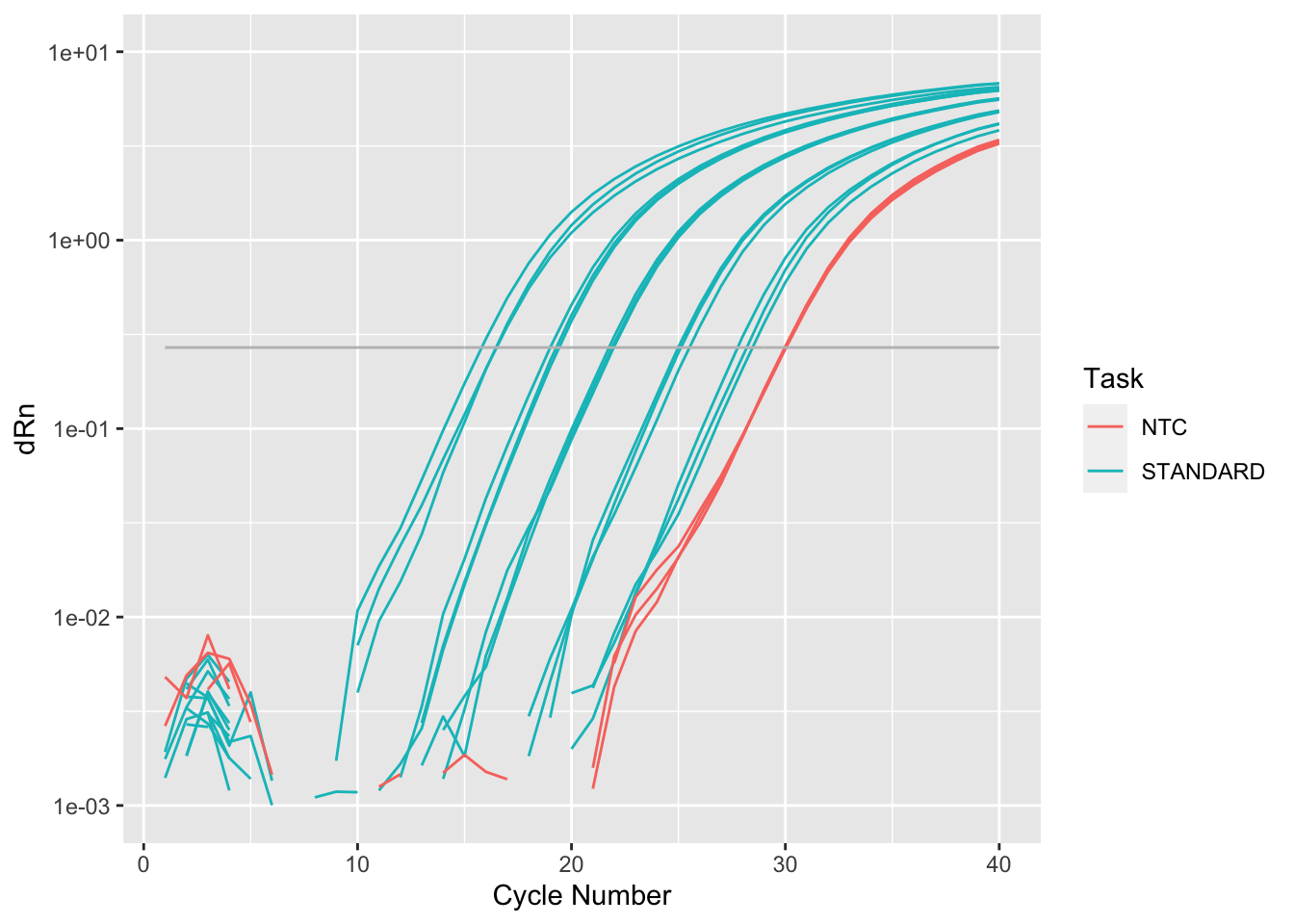
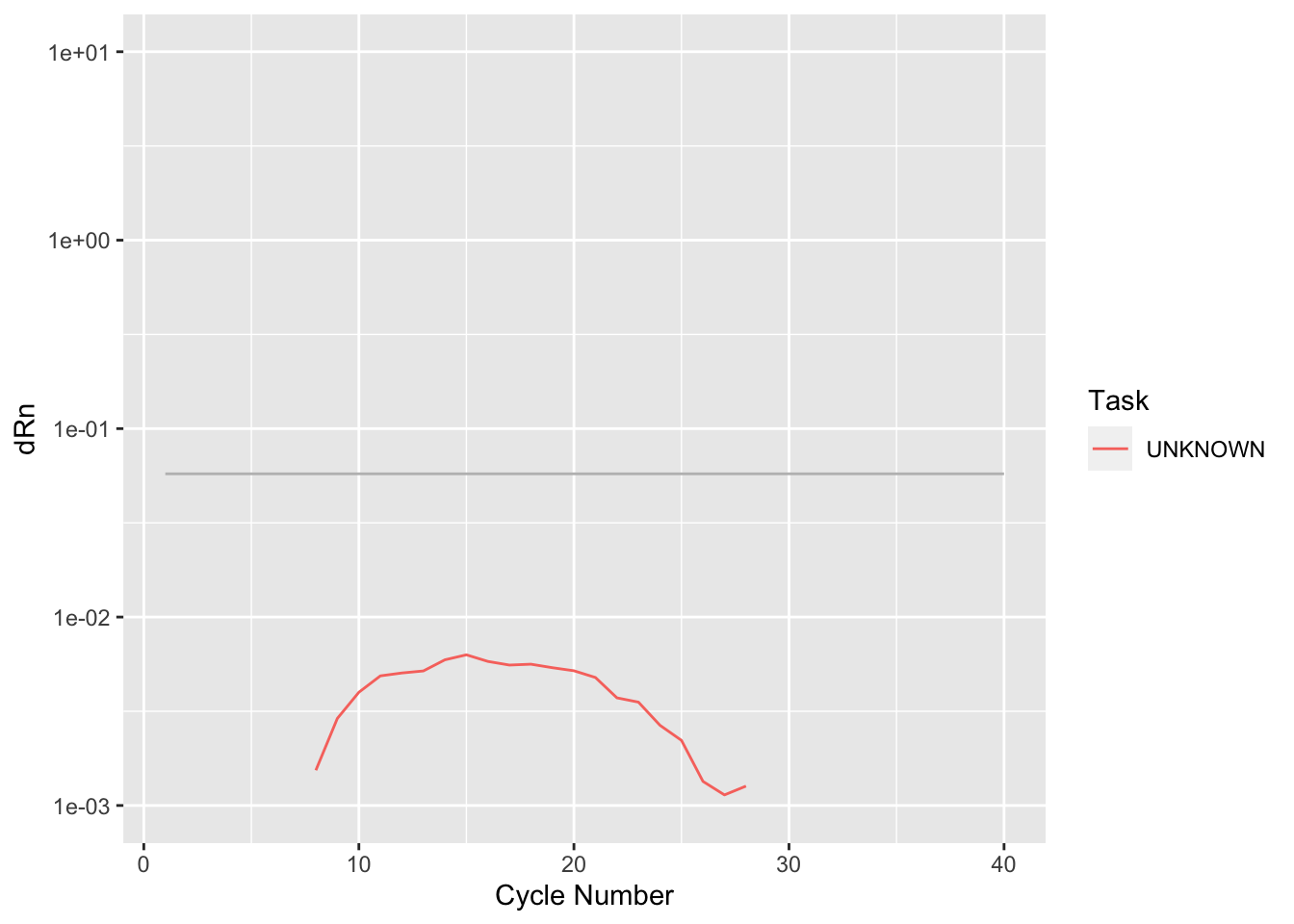
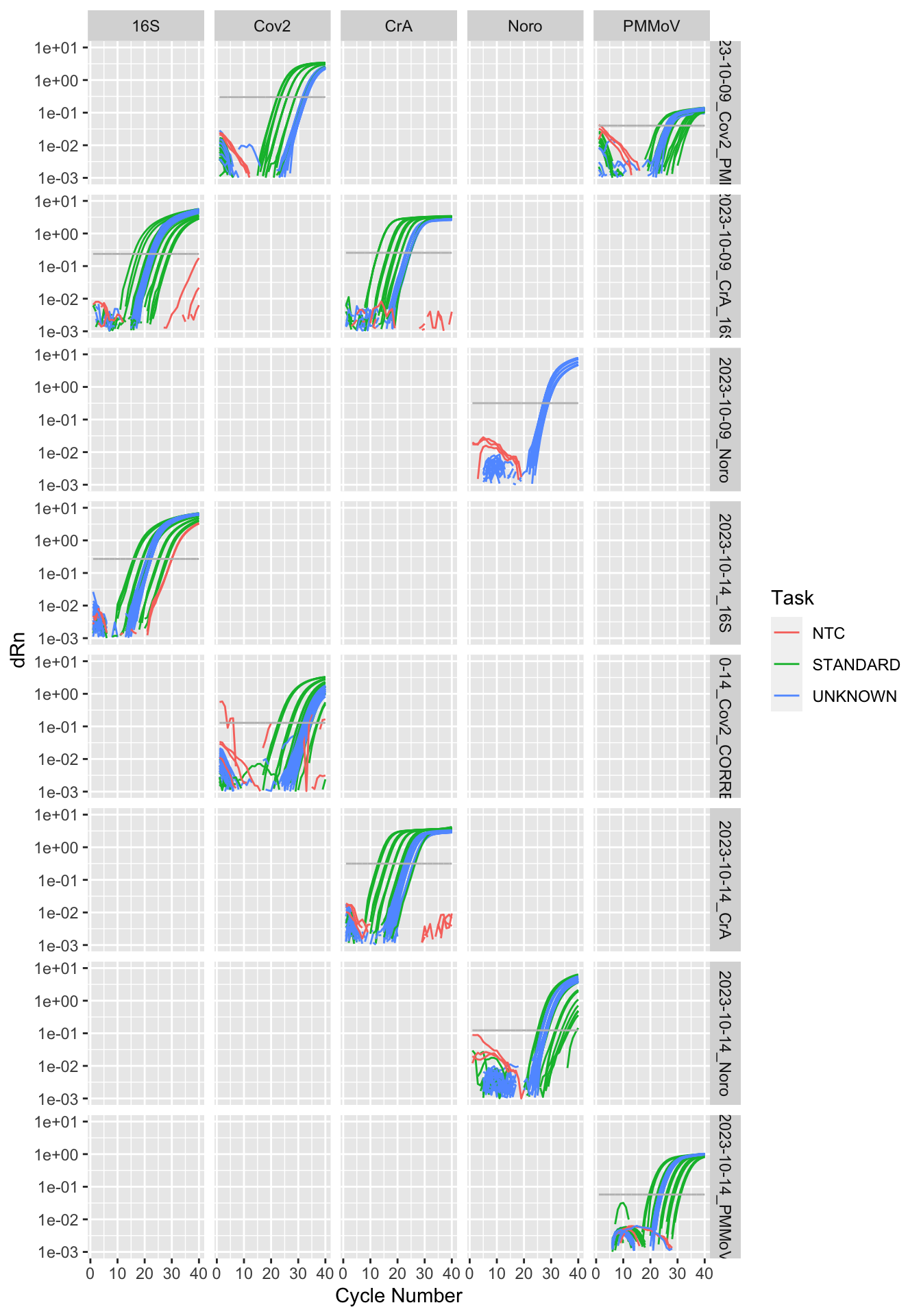
Rows: 19,240
Columns: 39
$ Well <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ `Well Position` <chr> "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1"…
$ `Cycle Number` <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,…
$ Target <chr> "Cov2", "Cov2", "Cov2", "Cov2", "Cov2", "Cov2"…
$ Rn <dbl> 0.6443956, 0.6382574, 0.6284555, 0.6179680, 0.…
$ dRn <dbl> 2.836028e-02, 2.374099e-02, 1.545804e-02, 6.48…
$ Sample <chr> "1A", "1A", "1A", "1A", "1A", "1A", "1A", "1A"…
$ Omit <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALS…
$ plate <chr> "2023-10-09_Cov2_PMMV", "2023-10-09_Cov2_PMMV"…
$ well_row <chr> "A", "A", "A", "A", "A", "A", "A", "A", "A", "…
$ well_col <chr> "1", "1", "1", "1", "1", "1", "1", "1", "1", "…
$ Task <chr> "UNKNOWN", "UNKNOWN", "UNKNOWN", "UNKNOWN", "U…
$ Reporter <chr> "FAM", "FAM", "FAM", "FAM", "FAM", "FAM", "FAM…
$ Quencher <chr> "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "N…
$ `Amp Status` <chr> "AMP", "AMP", "AMP", "AMP", "AMP", "AMP", "AMP…
$ `Amp Score` <dbl> 1.391558, 1.391558, 1.391558, 1.391558, 1.3915…
$ `Curve Quality` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ `Result Quality Issues` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ Cq <dbl> 33.15919, 33.15919, 33.15919, 33.15919, 33.159…
$ `Cq Confidence` <dbl> 0.9759085, 0.9759085, 0.9759085, 0.9759085, 0.…
$ `Cq Mean` <dbl> 32.93495, 32.93495, 32.93495, 32.93495, 32.934…
$ `Cq SD` <dbl> 0.2522912, 0.2522912, 0.2522912, 0.2522912, 0.…
$ `Auto Threshold` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ Threshold <dbl> 0.2999157, 0.2999157, 0.2999157, 0.2999157, 0.…
$ `Auto Baseline` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ `Baseline Start` <dbl> 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
$ `Baseline End` <dbl> 27, 27, 27, 27, 27, 27, 27, 27, 27, 27, 27, 27…
$ TreatmentGroup <chr> "Centrifuge + 0.45 filter", "Centrifuge + 0.45…
$ VortexMin <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ CFSpeed <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ CollectionDate <date> 2023-10-02, 2023-10-02, 2023-10-02, 2023-10-0…
$ Source <chr> "N-S mix", "N-S mix", "N-S mix", "N-S mix", "N…
$ Volume <dbl> 280, 280, 280, 280, 280, 280, 280, 280, 280, 2…
$ Qubit_ID <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ ProcessingHandler <chr> "Ari", "Ari", "Ari", "Ari", "Ari", "Ari", "Ari…
$ ExtractionHandler <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ `qPCR date` <date> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ qPCRHandler <chr> "Olivia", "Olivia", "Olivia", "Olivia", "Olivi…
$ qPCR_dilution <chr> "1:5", "1:5", "1:5", "1:5", "1:5", "1:5", "1:5…