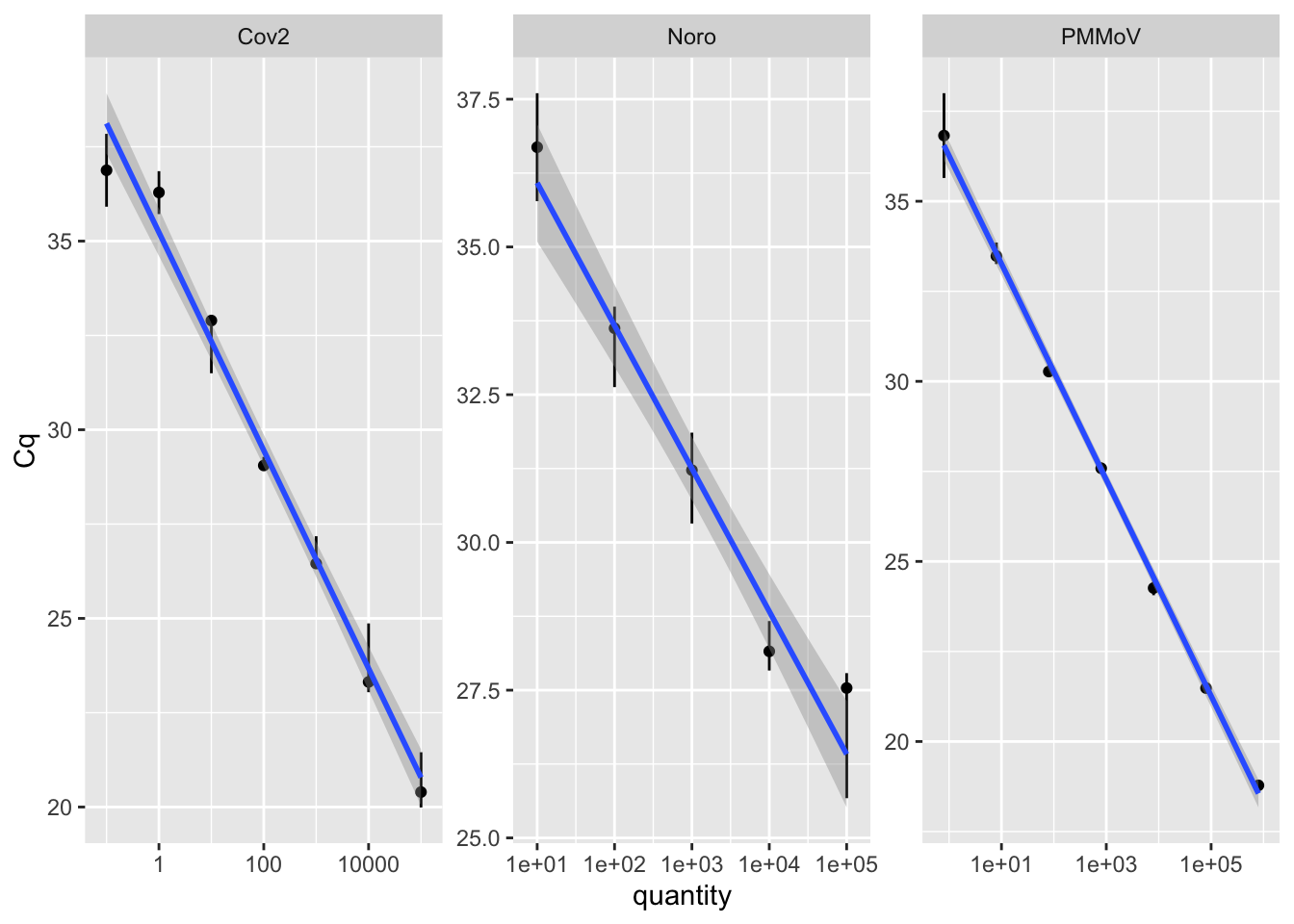
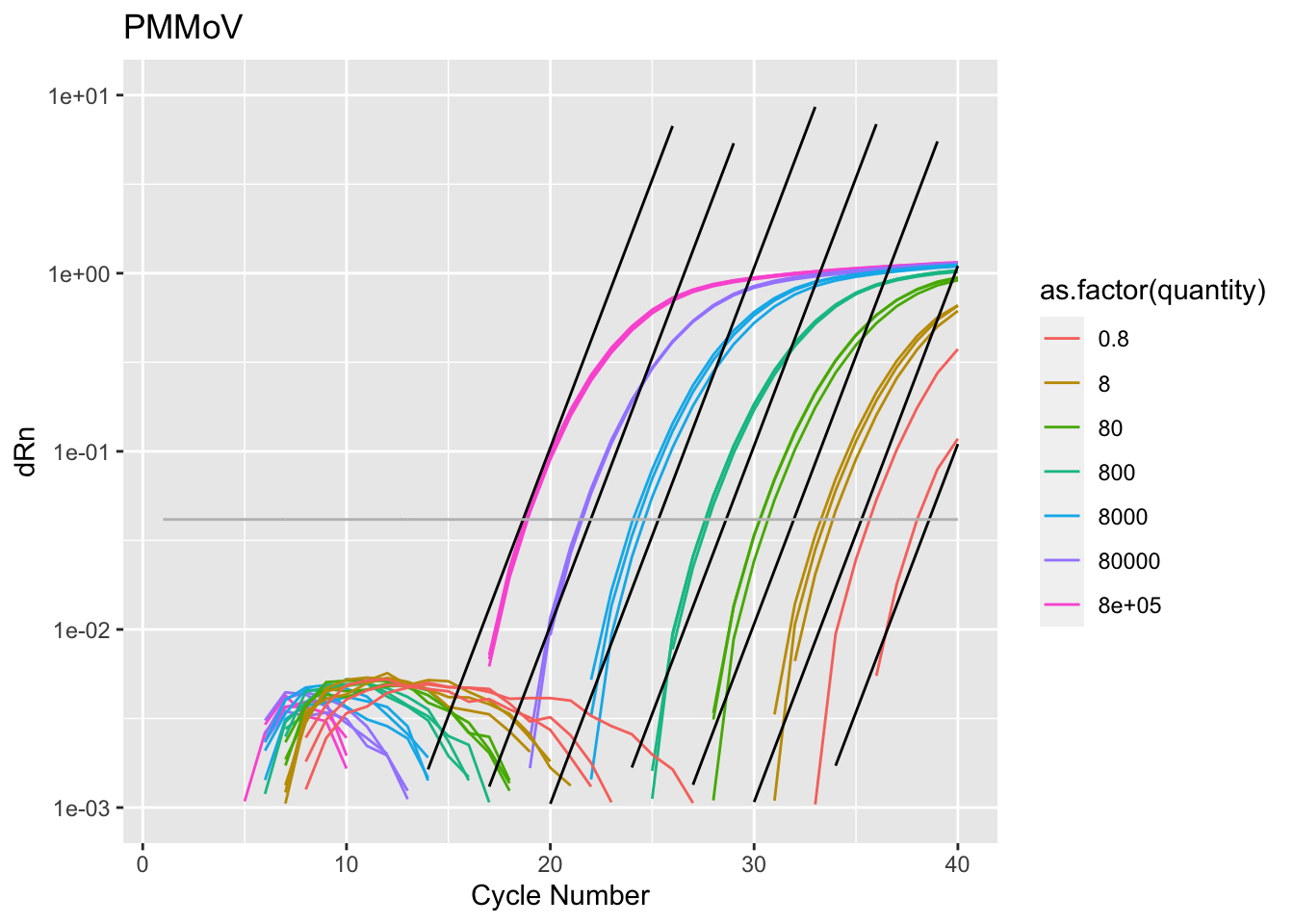
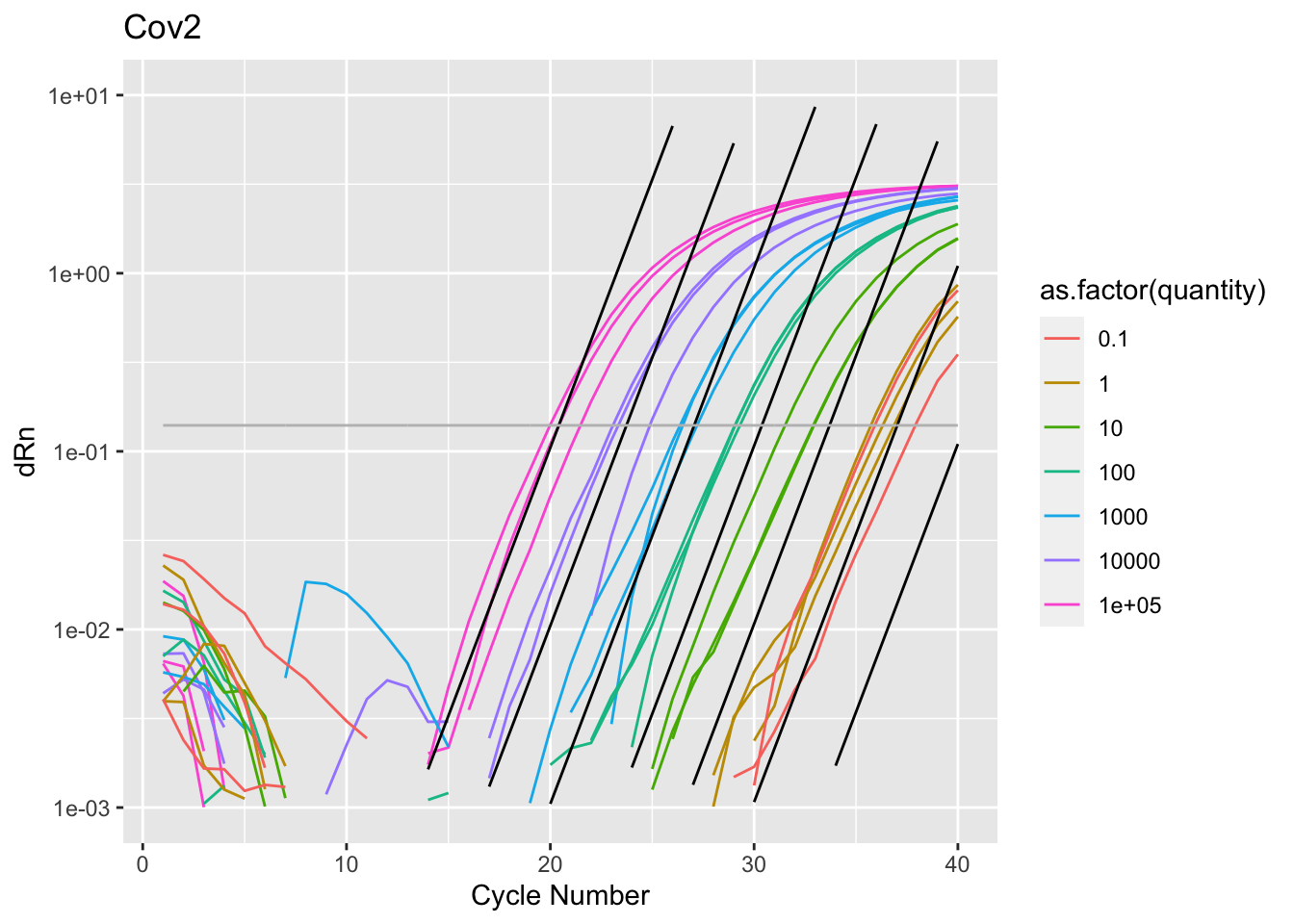
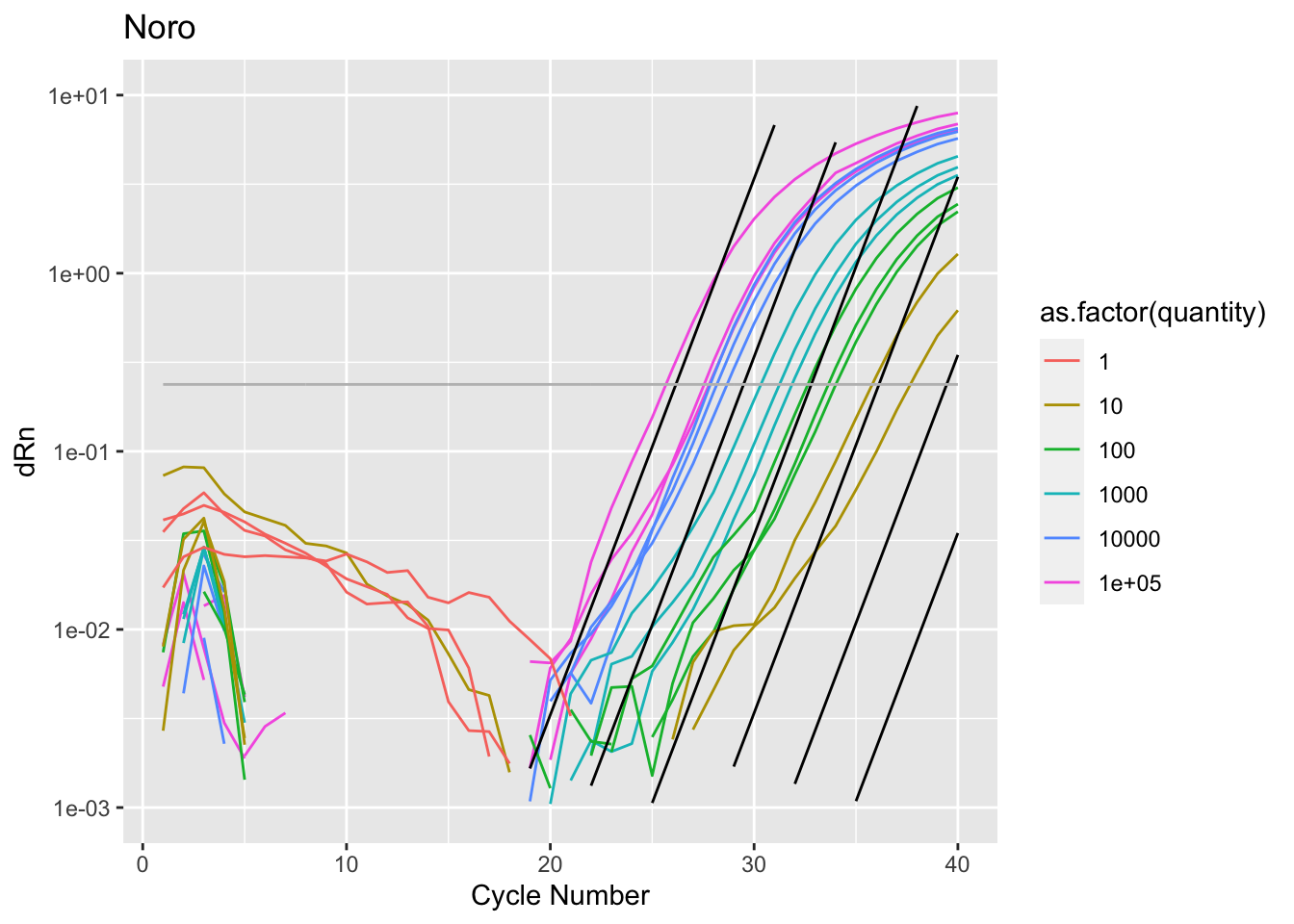
Rows: 2,760
Columns: 25
$ Well <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ `Well Position` <chr> "A1", "A1", "A1", "A1", "A1", "A1", "A1", "A1"…
$ `Cycle Number` <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,…
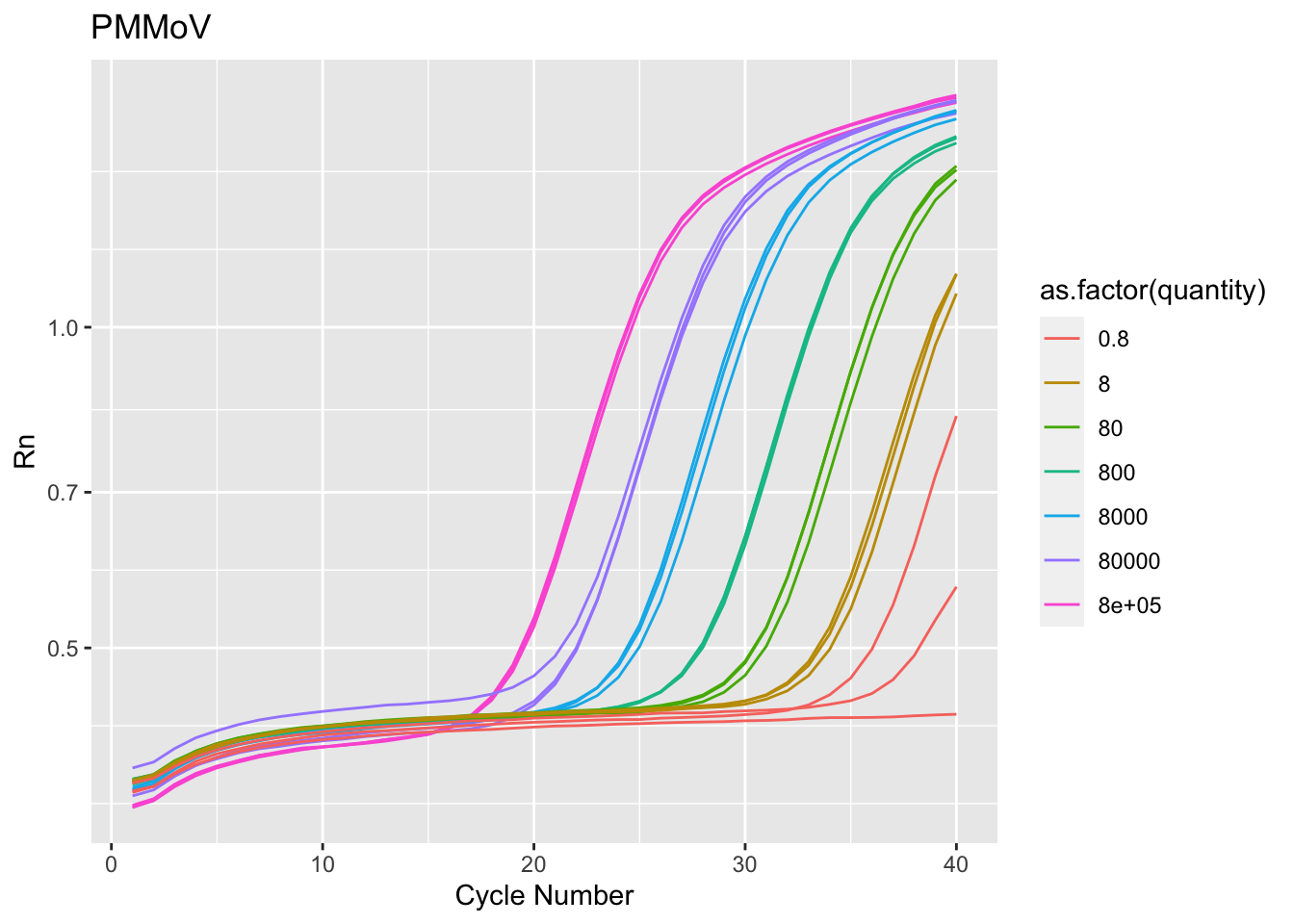
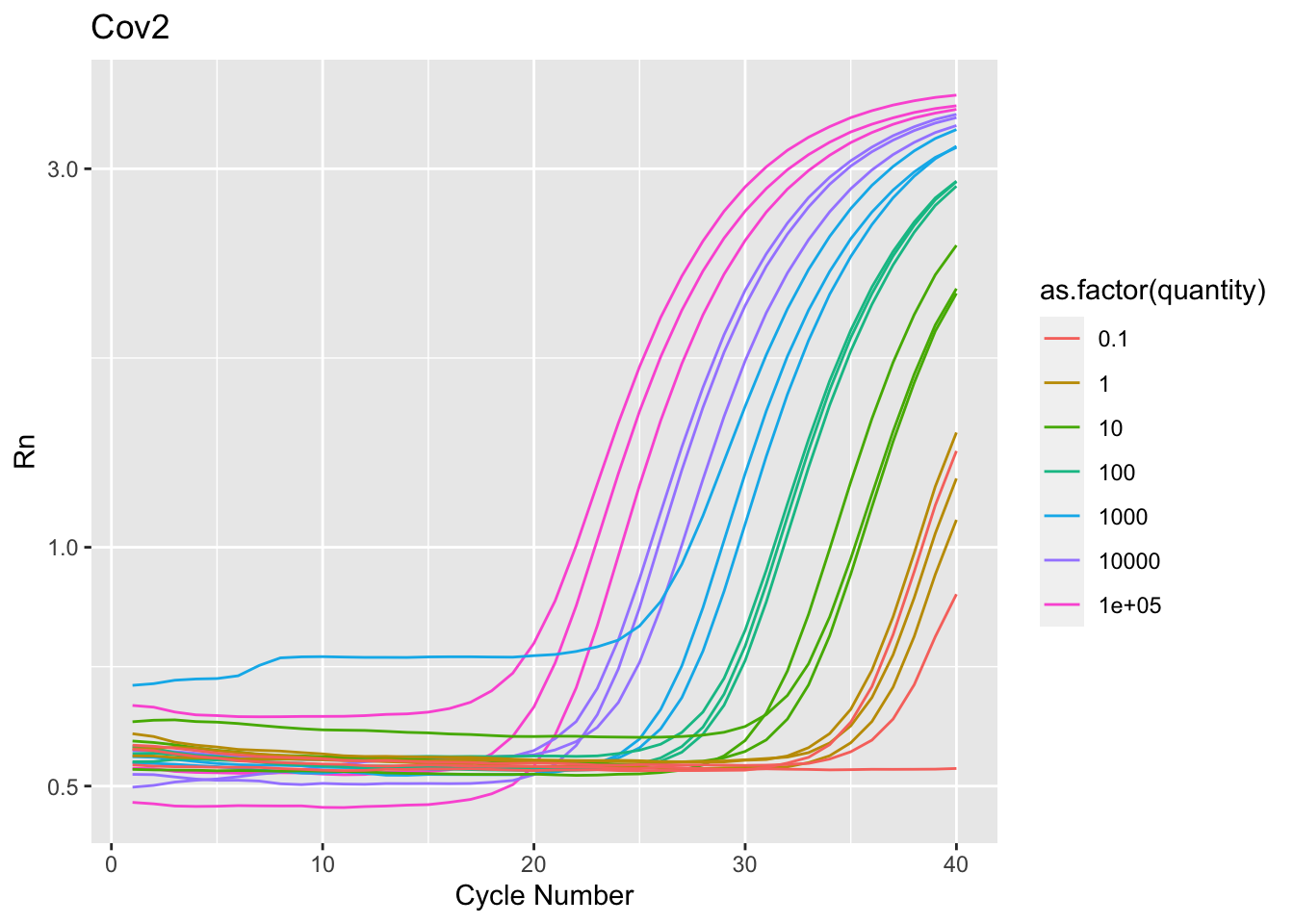
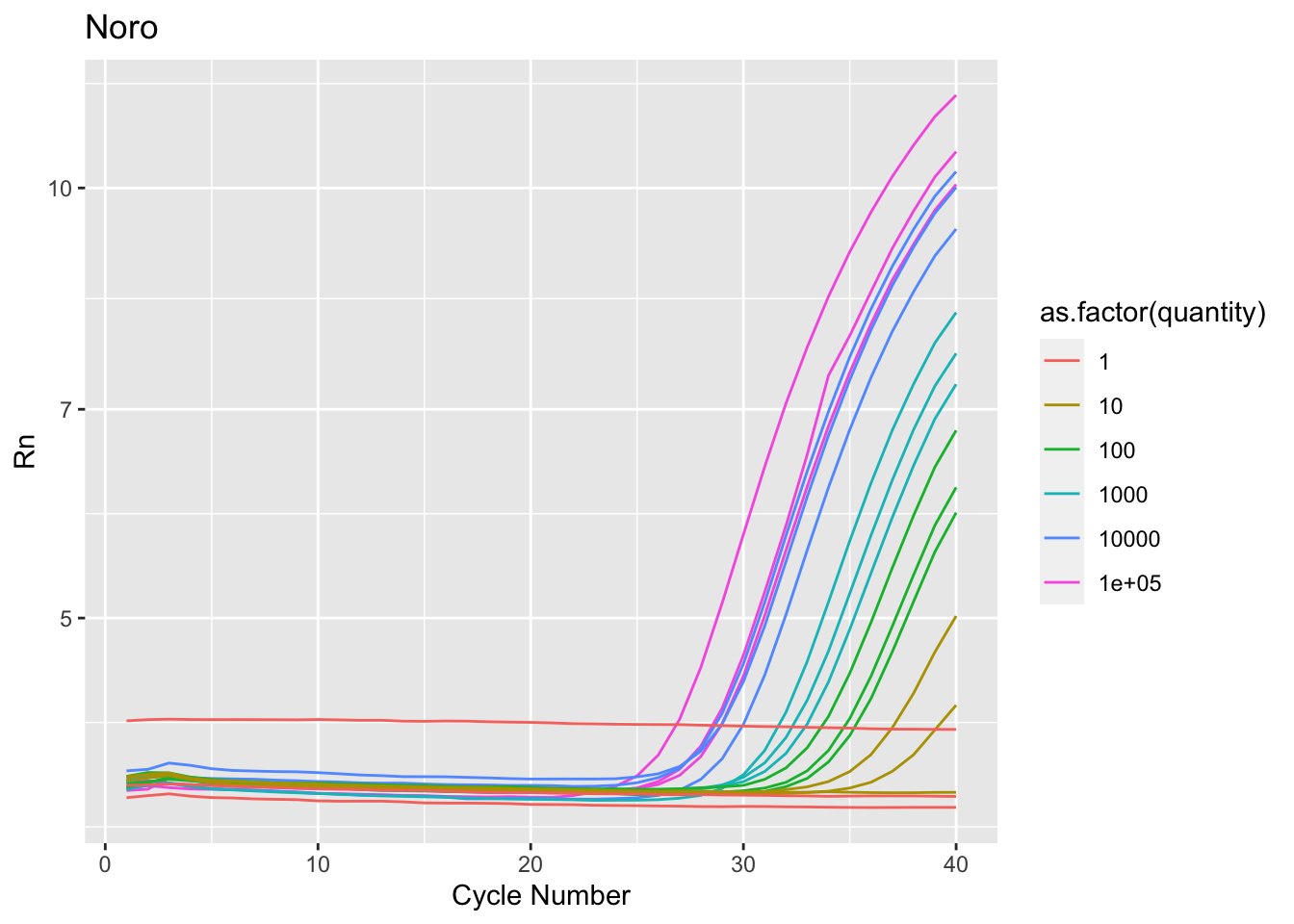
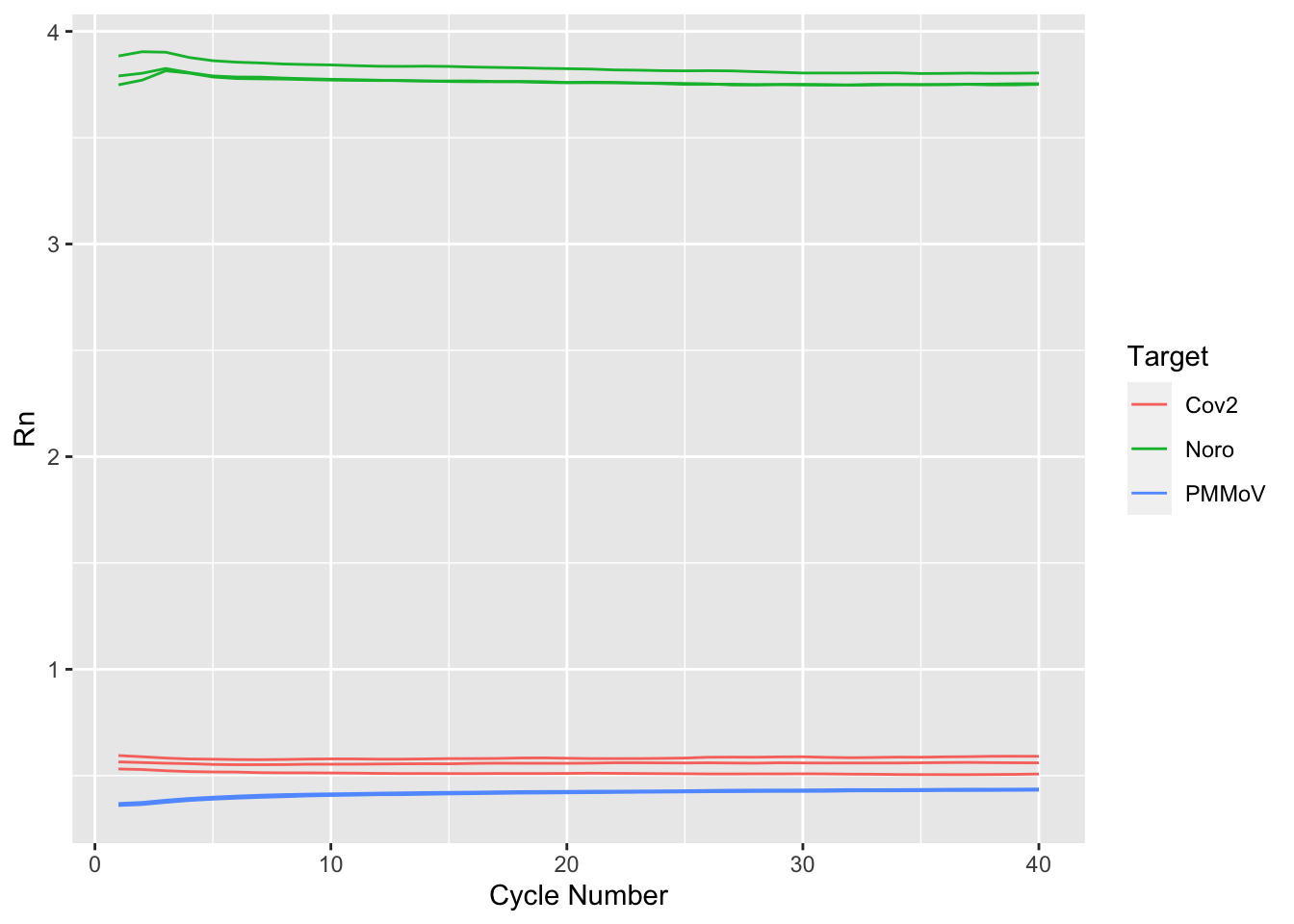
$ Target <chr> "Cov2", "Cov2", "Cov2", "Cov2", "Cov2", "Cov2"…
$ Rn <dbl> 0.6318336, 0.6287087, 0.6199436, 0.6148189, 0.…
$ dRn <dbl> 0.0186775648, 0.0154212920, 0.0065248104, 0.00…
$ Sample <dbl> 1e+05, 1e+05, 1e+05, 1e+05, 1e+05, 1e+05, 1e+0…
$ Omit <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALS…
$ Task <chr> "STANDARD", "STANDARD", "STANDARD", "STANDARD"…
$ Reporter <chr> "FAM", "FAM", "FAM", "FAM", "FAM", "FAM", "FAM…
$ Quencher <chr> "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "NFQ-MGB", "N…
$ `Amp Status` <chr> "AMP", "AMP", "AMP", "AMP", "AMP", "AMP", "AMP…
$ `Amp Score` <dbl> 1.336953, 1.336953, 1.336953, 1.336953, 1.3369…
$ `Curve Quality` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ `Result Quality Issues` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ Cq <dbl> 19.98854, 19.98854, 19.98854, 19.98854, 19.988…
$ `Cq Confidence` <dbl> 0.9907778, 0.9907778, 0.9907778, 0.9907778, 0.…
$ `Cq Mean` <dbl> 20.61243, 20.61243, 20.61243, 20.61243, 20.612…
$ `Cq SD` <dbl> 0.755257, 0.755257, 0.755257, 0.755257, 0.7552…
$ `Auto Threshold` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ Threshold <dbl> 0.1399036, 0.1399036, 0.1399036, 0.1399036, 0.…
$ `Auto Baseline` <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE…
$ `Baseline Start` <dbl> 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
$ `Baseline End` <dbl> 15, 15, 15, 15, 15, 15, 15, 15, 15, 15, 15, 15…
$ quantity <dbl> 1e+05, 1e+05, 1e+05, 1e+05, 1e+05, 1e+05, 1e+0…