Rows: 14 Columns: 17
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (8): Run ID, Assay Name, Test Name, Test Date, Qubit tube conc. units, O...
dbl (7): Qubit tube conc., Original sample conc., Sample Volume (uL), Diluti...
lgl (2): Std 3 RFU, Green RFU
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Rows: 14
Columns: 17
$ `Run ID` <chr> "2023-09-11_165152", "2023-09-11_165152"…
$ `Assay Name` <chr> "RNA High Sensitivity", "RNA High Sensit…
$ `Test Name` <chr> "Sample_#230911-165447", "Sample_#230911…
$ `Test Date` <chr> "09/11/2023 04:54:47 PM", "09/11/2023 04…
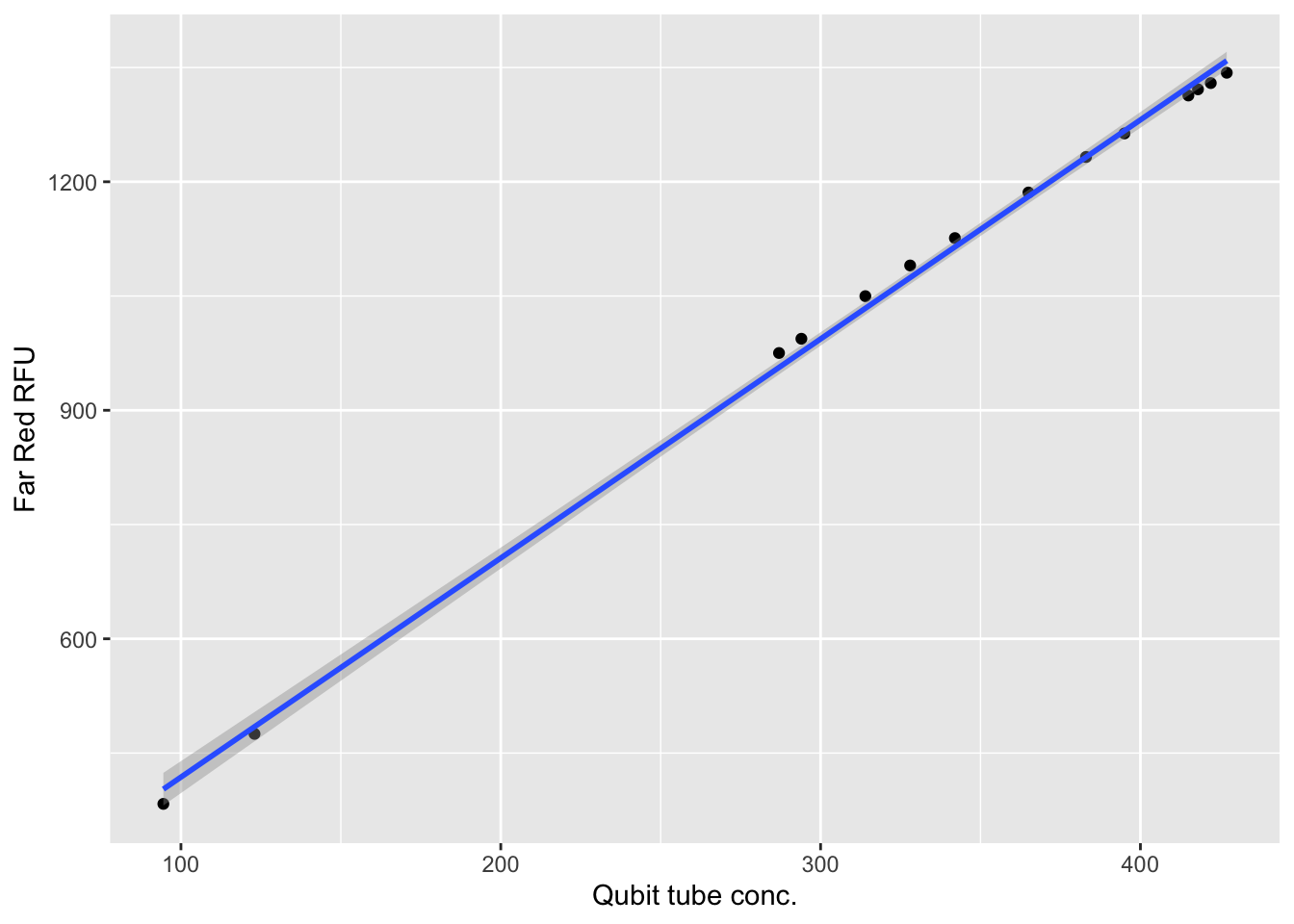
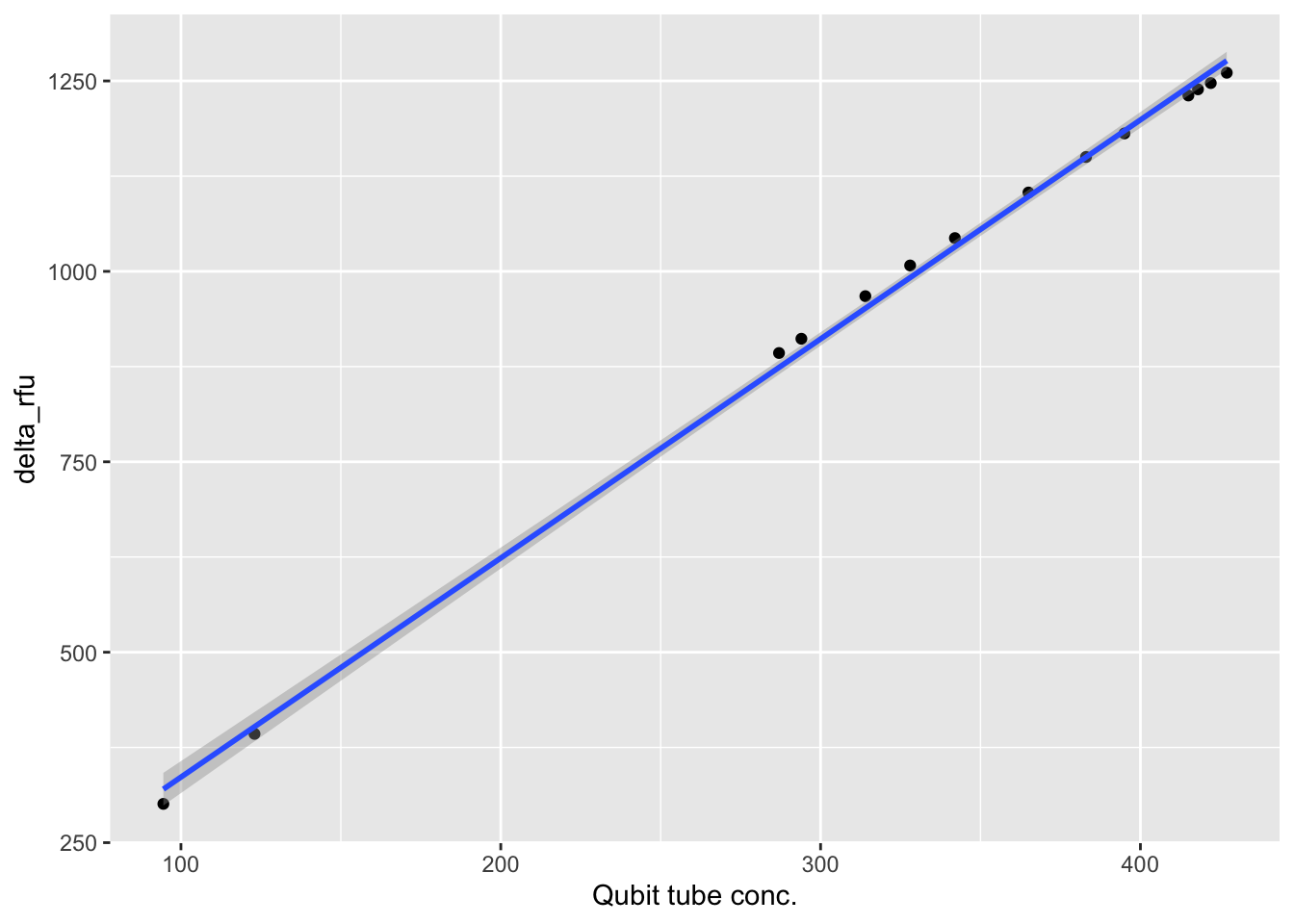
$ `Qubit tube conc.` <dbl> 123.0, 422.0, 415.0, 294.0, 287.0, 328.0…
$ `Qubit tube conc. units` <chr> "ng/mL", "ng/mL", "ng/mL", "ng/mL", "ng/…
$ `Original sample conc.` <dbl> 4.92, 16.90, 16.60, 11.80, 11.50, 13.10,…
$ `Original sample conc. units` <chr> "ng/uL", "ng/uL", "ng/uL", "ng/uL", "ng/…
$ `Sample Volume (uL)` <dbl> 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5
$ `Dilution Factor` <dbl> 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, …
$ `Std 1 RFU` <dbl> 82.37, 82.37, 82.37, 82.37, 82.37, 82.37…
$ `Std 2 RFU` <dbl> 1514.1, 1514.1, 1514.1, 1514.1, 1514.1, …
$ `Std 3 RFU` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ Excitation <chr> "Red", "Red", "Red", "Red", "Red", "Red"…
$ Emission <chr> "Far Red", "Far Red", "Far Red", "Far Re…
$ `Green RFU` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Far Red RFU` <dbl> 475.18, 1329.59, 1313.26, 993.90, 975.18…