Objectives
NOTE: Need sample metadata. I don’t know what the samples are.
- Test qPCR of PMMoV
- Test duplexed qPCR
- See Google Doc
Preliminary work
- Dan copied the
.eds file from the NAO qPCR data directory to the experiment directory and exported .csv files.
Data import
library(readr)
library(dplyr)
Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
library(purrr)
library(stringr)
library(ggplot2)
library(tidyr)
library(broom)
data_dir <-
"~/airport/[2023-09-13] PMMoV and Multiplex qPCR/"
filename_pattern <- "Results"
col_types <- list(
Target = col_character(),
Cq = col_double()
)
raw_data <- list.files(
paste0(data_dir, "qpcr"),
pattern = filename_pattern,
full.names = TRUE,
) |>
print() |>
map(function(f) {
read_csv(f,
skip = 23,
col_types = col_types,
)
}) |>
list_rbind()
[1] "/Users/dan/airport/[2023-09-13] PMMoV and Multiplex qPCR/qpcr/Ari PMMoV_Results_20230925_115306.csv"
Warning: One or more parsing issues, call `problems()` on your data frame for details,
e.g.:
dat <- vroom(...)
problems(dat)
# A tibble: 63 × 21
Well `Well Position` Omit Sample Target Task Reporter Quencher
<dbl> <chr> <lgl> <chr> <chr> <chr> <chr> <chr>
1 1 A1 FALSE 1 PPMoV UNKNOWN VIC NFQ-MGB
2 2 A2 FALSE 1 PPMoV UNKNOWN VIC NFQ-MGB
3 3 A3 FALSE 1 PPMoV UNKNOWN VIC NFQ-MGB
4 5 A5 FALSE 1 N2 UNKNOWN FAM NFQ-MGB
5 6 A6 FALSE 1 N2 UNKNOWN FAM NFQ-MGB
6 8 A8 FALSE NTC N2 UNKNOWN FAM NFQ-MGB
7 10 A10 FALSE 1 N2 UNKNOWN FAM NFQ-MGB
8 10 A10 FALSE 1 PPMoV UNKNOWN VIC NFQ-MGB
9 11 A11 FALSE 1 N2 UNKNOWN FAM NFQ-MGB
10 11 A11 FALSE 1 PPMoV UNKNOWN VIC NFQ-MGB
# ℹ 53 more rows
# ℹ 13 more variables: `Amp Status` <chr>, `Amp Score` <dbl>,
# `Curve Quality` <lgl>, `Result Quality Issues` <lgl>, Cq <dbl>,
# `Cq Confidence` <dbl>, `Cq Mean` <dbl>, `Cq SD` <dbl>,
# `Auto Threshold` <lgl>, Threshold <dbl>, `Auto Baseline` <lgl>,
# `Baseline Start` <dbl>, `Baseline End` <dbl>
It looks like the software splits duplexed targets into two rows, but doesn’t save the information about multiplexing.
raw_data |> count(Target, Reporter, Sample, Quencher)
# A tibble: 19 × 5
Target Reporter Sample Quencher n
<chr> <chr> <chr> <chr> <int>
1 N2 FAM 1 NFQ-MGB 5
2 N2 FAM 2 NFQ-MGB 5
3 N2 FAM NTC NFQ-MGB 2
4 N2 FAM SC1 NFQ-MGB 3
5 N2 FAM SC2 NFQ-MGB 3
6 N2 FAM SC3 NFQ-MGB 3
7 N2 FAM SC4 NFQ-MGB 3
8 N2 FAM SC5 NFQ-MGB 3
9 N2 FAM SC6 NFQ-MGB 3
10 N2 FAM <NA> NFQ-MGB 2
11 PPMoV VIC 1 NFQ-MGB 6
12 PPMoV VIC 2 NFQ-MGB 6
13 PPMoV VIC NTC NFQ-MGB 4
14 PPMoV VIC SC1 NFQ-MGB 1
15 PPMoV VIC SC2 NFQ-MGB 3
16 PPMoV VIC SC3 NFQ-MGB 3
17 PPMoV VIC SC4 NFQ-MGB 3
18 PPMoV VIC SC5 NFQ-MGB 3
19 PPMoV VIC <NA> NFQ-MGB 2
raw_data |>
count(`Well Position`) |>
print(n = Inf)
# A tibble: 54 × 2
`Well Position` n
<chr> <int>
1 A1 1
2 A10 2
3 A11 2
4 A12 2
5 A2 1
6 A3 1
7 A5 1
8 A6 1
9 A8 1
10 B1 1
11 B10 2
12 B11 2
13 B12 2
14 B2 1
15 B3 1
16 B5 1
17 B6 1
18 C1 1
19 C10 2
20 C11 2
21 C12 2
22 C2 1
23 C3 1
24 C5 1
25 C6 1
26 C7 1
27 D3 1
28 D5 1
29 D6 1
30 D7 1
31 E1 1
32 E2 1
33 E3 1
34 E5 1
35 E6 1
36 E7 1
37 F1 1
38 F2 1
39 F3 1
40 F5 1
41 F6 1
42 F7 1
43 G1 1
44 G2 1
45 G3 1
46 G5 1
47 G6 1
48 G7 1
49 H1 1
50 H2 1
51 H3 1
52 H5 1
53 H6 1
54 H7 1
We can count the multiplexing and save it as a column:
with_counts <- raw_data |>
left_join(count(raw_data, Well, name = "n_multiplex"), by = join_by(Well))
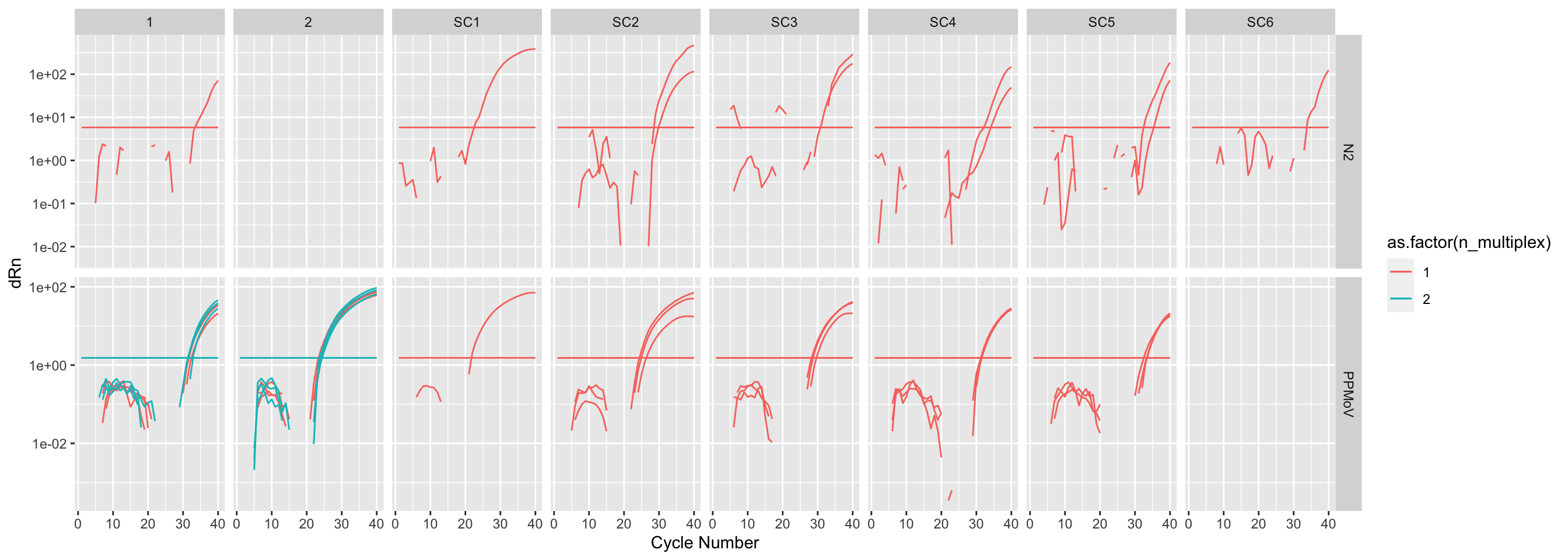
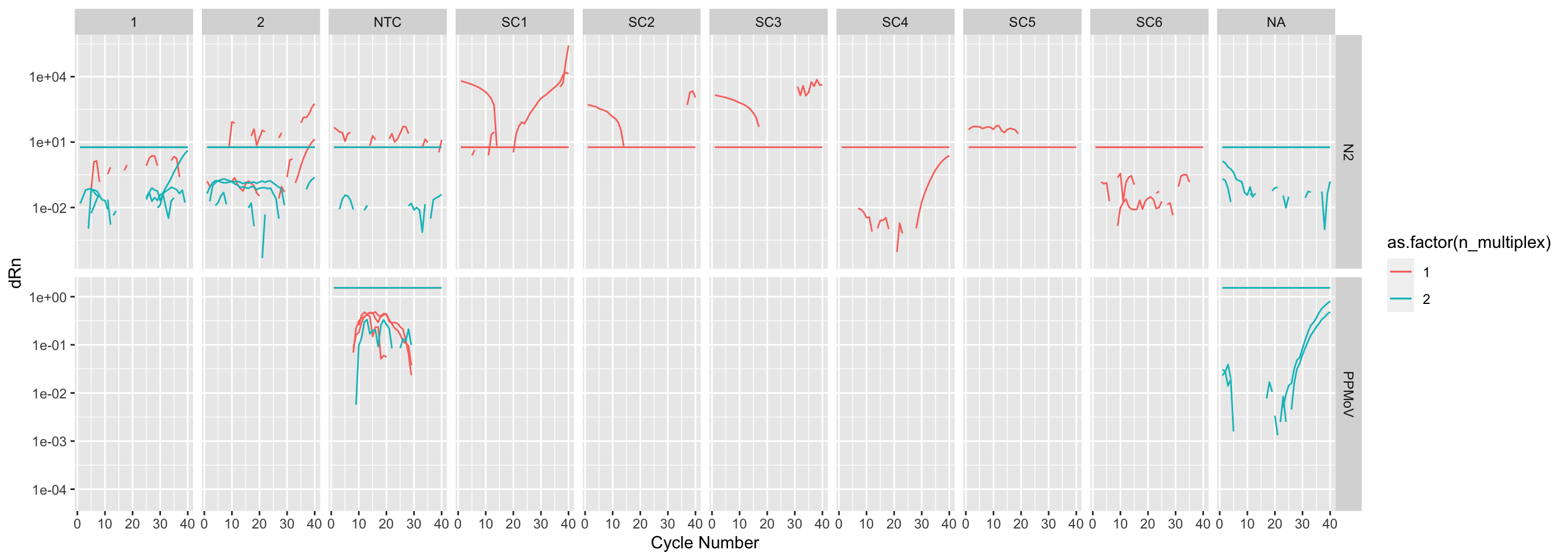
It looks like the multiplexed wells didn’t amplifiy well. No N2 wells amplified. It also looks like there was poor amp in general.
with_counts |>
count(Target, Reporter, Sample, n_multiplex, `Amp Status`) |>
print(n = Inf)
# A tibble: 32 × 6
Target Reporter Sample n_multiplex `Amp Status` n
<chr> <chr> <chr> <int> <chr> <int>
1 N2 FAM 1 1 AMP 1
2 N2 FAM 1 1 NO_AMP 1
3 N2 FAM 1 2 NO_AMP 3
4 N2 FAM 2 1 NO_AMP 2
5 N2 FAM 2 2 NO_AMP 3
6 N2 FAM NTC 1 NO_AMP 1
7 N2 FAM NTC 2 NO_AMP 1
8 N2 FAM SC1 1 AMP 1
9 N2 FAM SC1 1 NO_AMP 2
10 N2 FAM SC2 1 AMP 2
11 N2 FAM SC2 1 NO_AMP 1
12 N2 FAM SC3 1 AMP 2
13 N2 FAM SC3 1 NO_AMP 1
14 N2 FAM SC4 1 AMP 2
15 N2 FAM SC4 1 NO_AMP 1
16 N2 FAM SC5 1 AMP 2
17 N2 FAM SC5 1 NO_AMP 1
18 N2 FAM SC6 1 AMP 1
19 N2 FAM SC6 1 NO_AMP 2
20 N2 FAM <NA> 2 NO_AMP 2
21 PPMoV VIC 1 1 AMP 3
22 PPMoV VIC 1 2 AMP 3
23 PPMoV VIC 2 1 AMP 3
24 PPMoV VIC 2 2 AMP 3
25 PPMoV VIC NTC 1 NO_AMP 3
26 PPMoV VIC NTC 2 NO_AMP 1
27 PPMoV VIC SC1 1 AMP 1
28 PPMoV VIC SC2 1 AMP 3
29 PPMoV VIC SC3 1 AMP 3
30 PPMoV VIC SC4 1 AMP 3
31 PPMoV VIC SC5 1 AMP 3
32 PPMoV VIC <NA> 2 NO_AMP 2
with_counts |>
count(Target, n_multiplex, `Amp Status`) |>
print(n = Inf)
# A tibble: 7 × 4
Target n_multiplex `Amp Status` n
<chr> <int> <chr> <int>
1 N2 1 AMP 11
2 N2 1 NO_AMP 12
3 N2 2 NO_AMP 9
4 PPMoV 1 AMP 19
5 PPMoV 1 NO_AMP 3
6 PPMoV 2 AMP 6
7 PPMoV 2 NO_AMP 3
Check that the thresholds are the same for every well with the same target
with_counts |>
count(Target, Threshold) |>
print(n = Inf)
# A tibble: 2 × 3
Target Threshold n
<chr> <dbl> <int>
1 N2 5.82 32
2 PPMoV 1.53 31
Amplification curves
amp_data <- list.files(
paste0(data_dir, "qpcr"),
pattern = "Amplification Data",
full.names = TRUE,
) |>
map(function(f) {
read_csv(f,
skip = 23,
col_types = col_types,
)
}) |>
list_rbind() |>
left_join(with_counts,
by = join_by(Well, `Well Position`, Sample, Target)
) |>
print()
Warning: The following named parsers don't match the column names: Cq
# A tibble: 2,520 × 26
Well `Well Position` `Cycle Number` Target Rn dRn Sample Omit.x
<dbl> <chr> <dbl> <chr> <dbl> <dbl> <chr> <lgl>
1 1 A1 1 PPMoV 13.5 -1.41 1 FALSE
2 1 A1 2 PPMoV 13.8 -1.23 1 FALSE
3 1 A1 3 PPMoV 14.4 -0.752 1 FALSE
4 1 A1 4 PPMoV 14.8 -0.474 1 FALSE
5 1 A1 5 PPMoV 15.2 -0.290 1 FALSE
6 1 A1 6 PPMoV 15.5 -0.0864 1 FALSE
7 1 A1 7 PPMoV 15.8 0.0333 1 FALSE
8 1 A1 8 PPMoV 16.0 0.138 1 FALSE
9 1 A1 9 PPMoV 16.3 0.272 1 FALSE
10 1 A1 10 PPMoV 16.5 0.302 1 FALSE
# ℹ 2,510 more rows
# ℹ 18 more variables: Omit.y <lgl>, Task <chr>, Reporter <chr>,
# Quencher <chr>, `Amp Status` <chr>, `Amp Score` <dbl>,
# `Curve Quality` <lgl>, `Result Quality Issues` <lgl>, Cq <dbl>,
# `Cq Confidence` <dbl>, `Cq Mean` <dbl>, `Cq SD` <dbl>,
# `Auto Threshold` <lgl>, Threshold <dbl>, `Auto Baseline` <lgl>,
# `Baseline Start` <dbl>, `Baseline End` <dbl>, n_multiplex <int>
Amplification curves by sample
The one that amplified:
amp_data |>
filter(`Amp Status` == "AMP") |>
ggplot(mapping = aes(
x = `Cycle Number`,
y = dRn,
group = Well,
color = as.factor(n_multiplex),
)) +
geom_line() +
geom_line(mapping = aes(
x = `Cycle Number`,
y = Threshold,
)) +
scale_y_log10() +
facet_grid(
rows = vars(Target), cols = vars(Sample), scales = "free_y"
)
Warning in self$trans$transform(x): NaNs produced
Warning: Transformation introduced infinite values in continuous y-axis
Warning: Removed 169 rows containing missing values (`geom_line()`).
The ones that didn’t:
amp_data |>
filter(`Amp Status` == "NO_AMP") |>
ggplot(mapping = aes(
x = `Cycle Number`,
y = dRn,
group = Well,
color = as.factor(n_multiplex),
)) +
geom_line() +
geom_line(mapping = aes(
x = `Cycle Number`,
y = Threshold,
)) +
scale_y_log10() +
facet_grid(
rows = vars(Target), cols = vars(Sample), scales = "free_y"
)
Warning in self$trans$transform(x): NaNs produced
Warning: Transformation introduced infinite values in continuous y-axis
Warning: Removed 187 rows containing missing values (`geom_line()`).
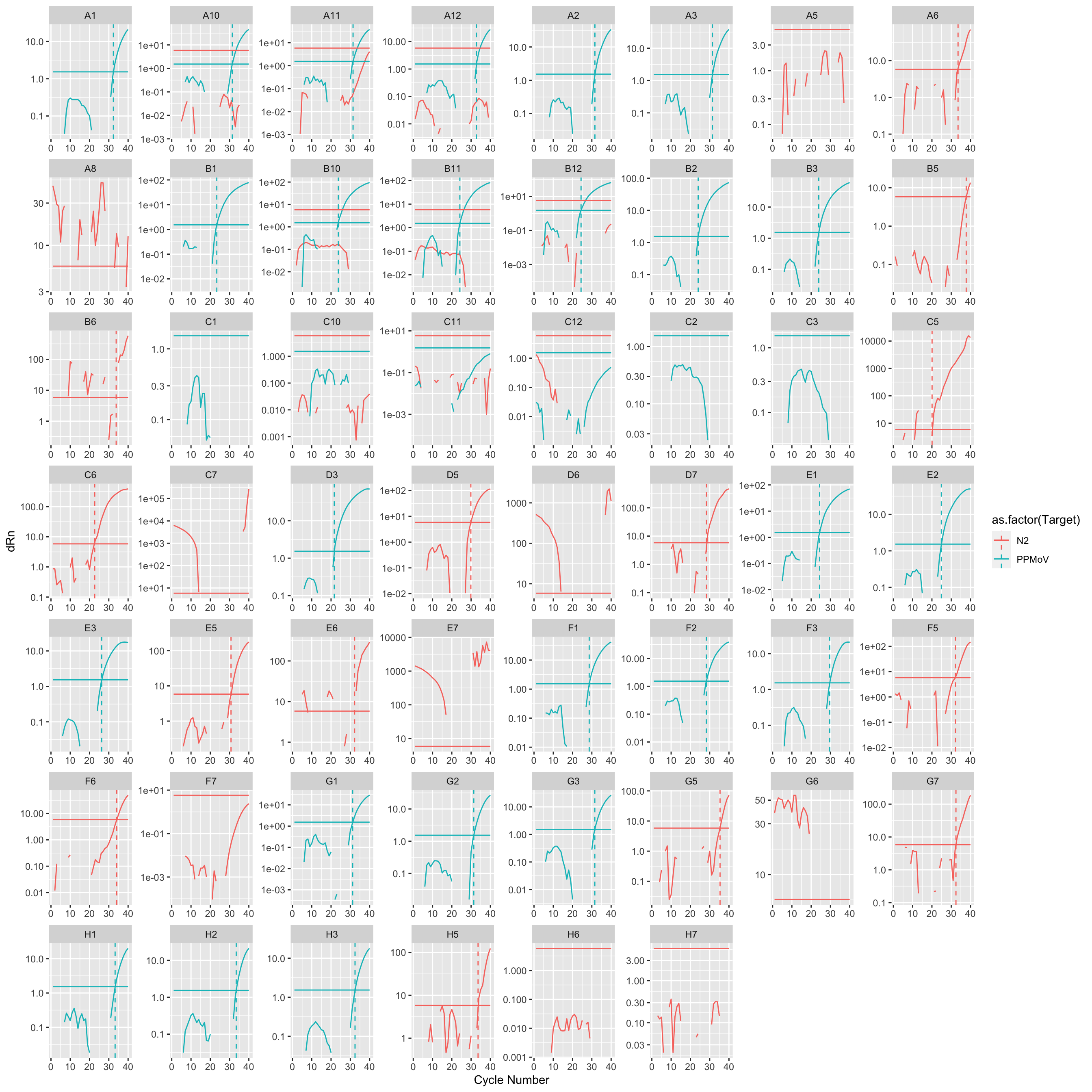
Amplification curves by well
amp_data |>
ggplot(mapping = aes(
x = `Cycle Number`,
y = dRn,
color = as.factor(Target),
)) +
geom_line() +
geom_line(mapping = aes(
x = `Cycle Number`,
y = Threshold
)) +
geom_vline(aes(xintercept = `Cq`, color = as.factor(Target)),
linetype = "dashed"
) +
scale_y_log10() +
facet_wrap(~`Well Position`, scales = "free")
Warning in self$trans$transform(x): NaNs produced
Warning: Transformation introduced infinite values in continuous y-axis
Warning: Removed 15 rows containing missing values (`geom_line()`).
Warning: Removed 960 rows containing missing values (`geom_vline()`).